Separate a 2 person mixture by ranking the possible contributor pairs.
rank_contributor_pairs(contrib_pairs, fit, max_rank = NULL)Arguments
- contrib_pairs
A
contrib_pairsobject obtained fromcontributor_pairs.- fit
A
disclapmixfitobject.- max_rank
Not used. Reserved for future use.
Value
A ranked_contrib_pairs object that is basically an order
vector and the probabilities for each pair (in the same order as given in
contrib_pairs), found by using fit. Note, that contributor
order is disregarded so that each contributor pair is only present once (and
not twice as would be the case if taking order into consideration).
See also
Examples
data(danes)
db <- as.matrix(danes[rep(1L:nrow(danes), danes$n), 1L:(ncol(danes) - 1L)])
set.seed(1)
true_contribs <- sample(1L:nrow(db), 2L)
h1 <- db[true_contribs[1L], ]
h2 <- db[true_contribs[2L], ]
db_ref <- db[-true_contribs, ]
h1h2 <- c(paste(h1, collapse = ";"), paste(h2, collapse = ";"))
tab_db <- table(apply(db, 1, paste, collapse = ";"))
tab_db_ref <- table(apply(db_ref, 1, paste, collapse = ";"))
tab_db[h1h2]
#>
#> 14;12;28;22;10;11;13;16;10;11 16;13;30;25;10;11;13;14;11;10
#> 9 1
tab_db_ref[h1h2]
#>
#> 14;12;28;22;10;11;13;16;10;11 <NA>
#> 8
rm(db) # To avoid use by accident
mixture <- generate_mixture(list(h1, h2))
possible_contributors <- contributor_pairs(mixture)
possible_contributors
#> Mixture:
#> DYS19 DYS389I DYS389II DYS390 DYS391 DYS392 DYS393 DYS437 DYS438 DYS439
#> 1 14,16 12,13 28,30 22,25 10 11 13 14,16 10,11 10,11
#>
#> Number of possible contributor pairs = 64
fits <- lapply(1L:5L, function(clus) disclapmix(db_ref, clusters = clus))
best_fit_BIC <- fits[[which.min(sapply(fits, function(fit) fit$BIC_marginal))]]
best_fit_BIC
#> disclapmixfit from 183 observations on 10 loci with 4 clusters.
#>
#> EM converged: TRUE
#> Number of central haplotype changes: 0
#> Total number of EM iterations: 16
#> Model observations (n*loci*clusters): 1830
#> Model parameters ((clusters*loci)+(loci+clusters-1)+(clusters-1)): 56
#> GLM method: internal_coef
#> Initial central haplotypes supplied: FALSE
#> Method to find initial central haplotypes: pam
ranked_contributors_BIC <- rank_contributor_pairs(possible_contributors, best_fit_BIC)
ranked_contributors_BIC
#> Mixture:
#> DYS19 DYS389I DYS389II DYS390 DYS391 DYS392 DYS393 DYS437 DYS438 DYS439
#> 1 14,16 12,13 28,30 22,25 10 11 13 14,16 10,11 10,11
#>
#> Contributor pairs = 64
#>
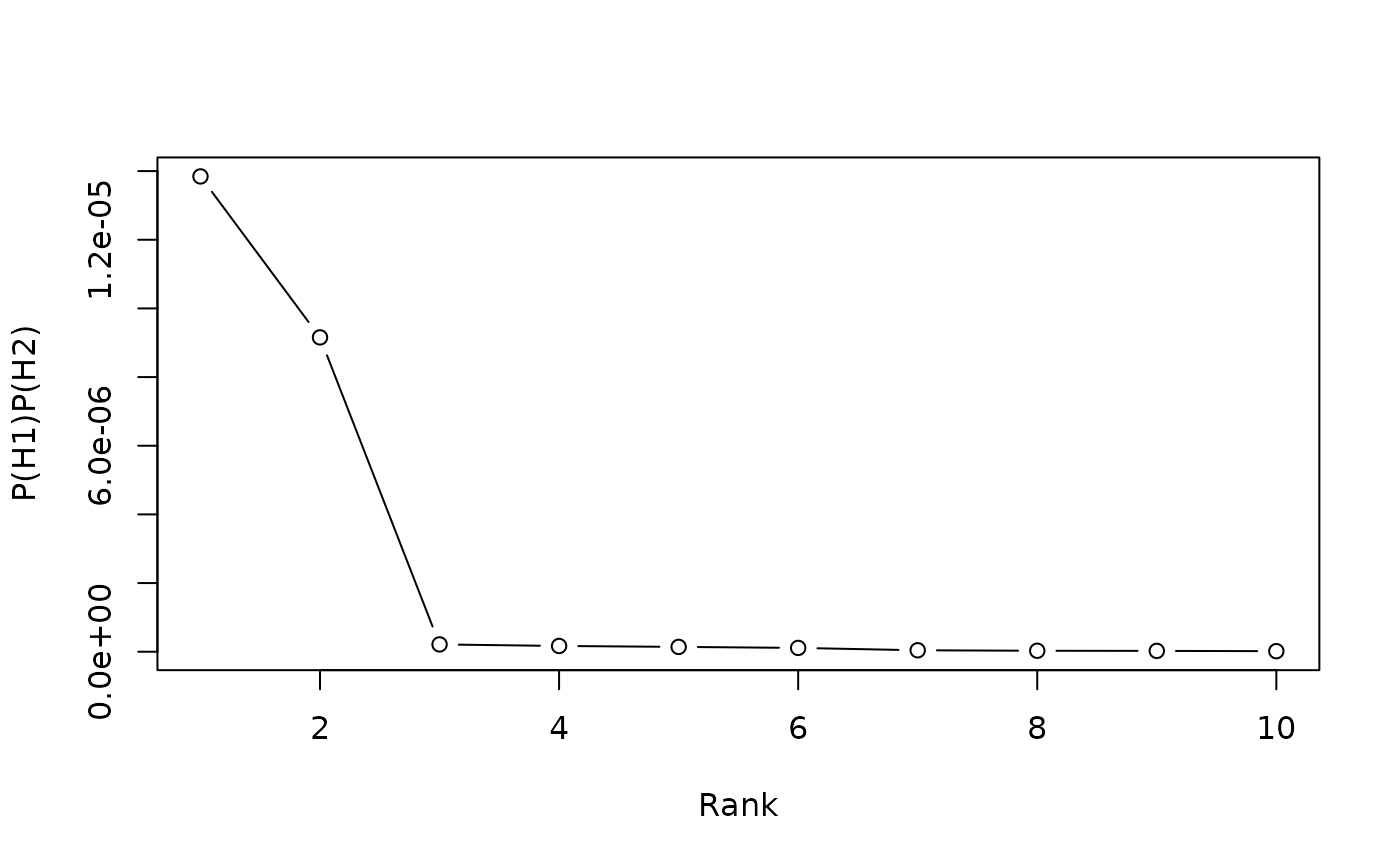
#> Sum of all (product of contributor pair haplotypes) = 2.375477e-05
#>
#> Showing rank 1-5:
#>
#> Rank 1 [ P(H1)*P(H2) = 1.384459e-05 ]:
#> DYS19 DYS389I DYS389II DYS390 DYS391 DYS392 DYS393 DYS437 DYS438 DYS439
#> H1 14 12 28 22 . . . 16 10 11
#> H2 16 13 30 25 . . . 14 11 10
#> Prob
#> H1 0.042522055
#> H2 0.000325586
#>
#> Rank 2 [ P(H1)*P(H2) = 9.155864e-06 ]:
#> DYS19 DYS389I DYS389II DYS390 DYS391 DYS392 DYS393 DYS437 DYS438 DYS439
#> H1 14 12 28 22 . . . 16 10 10
#> H2 16 13 30 25 . . . 14 11 11
#> Prob
#> H1 0.006408721
#> H2 0.001428657
#>
#> Rank 3 [ P(H1)*P(H2) = 2.128967e-07 ]:
#> DYS19 DYS389I DYS389II DYS390 DYS391 DYS392 DYS393 DYS437 DYS438 DYS439
#> H1 14 13 30 25 . . . 14 11 10
#> H2 16 12 28 22 . . . 16 10 11
#> Prob
#> H1 0.0003255861
#> H2 0.0006538875
#>
#> Rank 4 [ P(H1)*P(H2) = 1.67922e-07 ]:
#> DYS19 DYS389I DYS389II DYS390 DYS391 DYS392 DYS393 DYS437 DYS438 DYS439
#> H1 14 13 28 22 . . . 16 10 11
#> H2 16 12 30 25 . . . 14 11 10
#> Prob
#> H1 3.808357e-03
#> H2 4.409302e-05
#>
#> Rank 5 [ P(H1)*P(H2) = 1.407953e-07 ]:
#> DYS19 DYS389I DYS389II DYS390 DYS391 DYS392 DYS393 DYS437 DYS438 DYS439
#> H1 14 13 30 25 . . . 14 11 11
#> H2 16 12 28 22 . . . 16 10 10
#> Prob
#> H1 1.428657e-03
#> H2 9.855081e-05
#>
#> (59 contributor pairs hidden.)
plot(ranked_contributors_BIC, top = 10L, type = "b")
 get_rank(ranked_contributors_BIC, h1)
#> [1] 1
get_rank(ranked_contributors_BIC, h1)
#> [1] 1