For reproducibility:
set.seed(1)Data
library(disclapmix)
data(danes)
db <- as.matrix(danes[rep(1:nrow(danes), danes$n), 1:(ncol(danes)-1)])
str(db)
#> int [1:185, 1:10] 13 13 13 13 13 13 14 14 14 14 ...
#> - attr(*, "dimnames")=List of 2
#> ..$ : chr [1:185] "1" "2" "3" "4" ...
#> ..$ : chr [1:10] "DYS19" "DYS389I" "DYS389II" "DYS390" ...Using default parameters: partition around medoids (PAM)
Using partition around medoids (PAM) cluster method to find initial clusters:
default_fits <- disclapmix_adaptive(db, label = "PAM", margin = 5L)The label argument is added to the resulting fits (the
advantage is demonstrated later).
Using custom init_y_method: clustering large
applications (CLARA)
clara_fits <- disclapmix_adaptive(db, label = "CLARA", margin = 5L, init_y_method = "clara")Using custom init_y
Note the argument init_y_generator for
disclapmix_adaptive():
# Random observations:
my_init_y_generator <- function(k) {
# Or cluster::pam(), cluster::clara() or something else
db[sample(seq_len(nrow(db)), k, replace = FALSE), , drop = FALSE]
}
my_init_y_generator(1)
#> DYS19 DYS389I DYS389II DYS390 DYS391 DYS392 DYS393 DYS437 DYS438 DYS439
#> 133 16 12 28 24 10 11 12 16 9 12
my_init_y_generator(2)
#> DYS19 DYS389I DYS389II DYS390 DYS391 DYS392 DYS393 DYS437 DYS438 DYS439
#> 59 14 12 28 22 10 11 15 16 10 11
#> 48 14 13 29 24 10 15 13 15 12 12
custom_fits <- disclapmix_adaptive(db, label = "Custom",
margin = 1L, # Just demonstrating my_init_y_generator()
init_y_generator = my_init_y_generator)
rm(custom_fits_best) # To avoid using it by accident later
#> Warning in rm(custom_fits_best): object 'custom_fits_best' not foundNow, we can do multiple and take the best:
set.seed(2) # For reproducibility
custom_fits_extra <- replicate(5,
disclapmix_adaptive(db,
label = "Custom",
margin = 5L,
init_y_generator = my_init_y_generator,
# Random starting points may need more iterations
glm_control_maxit = 100L)
)
str(custom_fits_extra, 2)
#> List of 5
#> $ :List of 8
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> $ :List of 9
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> $ :List of 9
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> $ :List of 8
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> $ :List of 8
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
#> ..$ :List of 34
#> .. ..- attr(*, "class")= chr "disclapmixfit"
custom_fits_max_n <- max(sapply(custom_fits_extra, length))
custom_fits_best <- vector("list", custom_fits_max_n)
for (i in seq_len(custom_fits_max_n)) {
best_fit_i <- NULL
for (j in seq_along(custom_fits_extra)) {
if (length(custom_fits_extra[[j]]) < i) {
next
}
if (is.null(best_fit_i) ||
best_fit_i$BIC_marginal > custom_fits_extra[[j]][[i]]$BIC_marginal) {
best_fit_i <- custom_fits_extra[[j]][[i]]
}
}
custom_fits_best[[i]] <- best_fit_i
}Visualising
First we put all fits into a single list:
fits <- c(default_fits, clara_fits, custom_fits_best)And then construct a data frame with summary results:
d <- data.frame(
Label = sapply(fits, function(x) x$label),
BIC = sapply(fits, function(x) x$BIC_marginal),
Clusters = sapply(fits, function(x) nrow(x$y))
)
library(ggplot2)
ggplot(d, aes(Clusters, BIC, color = Label)) +
geom_point() +
geom_line() +
scale_x_continuous(breaks = unique(d$Clusters)) +
theme_bw()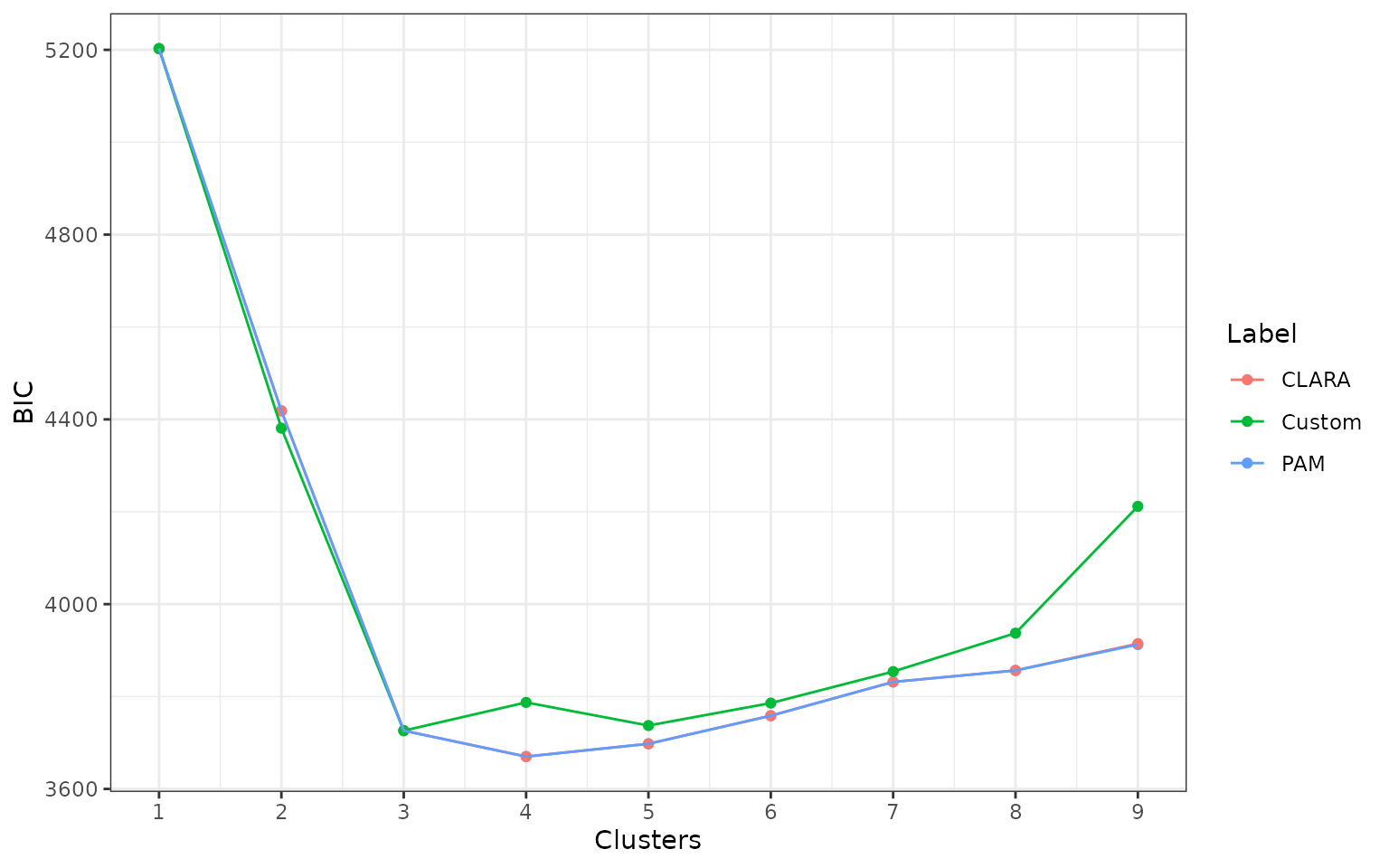
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
d %>%
group_by(Label) %>%
summarise(best_clusters = Clusters[which.min(BIC)])
#> # A tibble: 3 × 2
#> Label best_clusters
#> <chr> <int>
#> 1 CLARA 4
#> 2 Custom 3
#> 3 PAM 4Multiple criteria
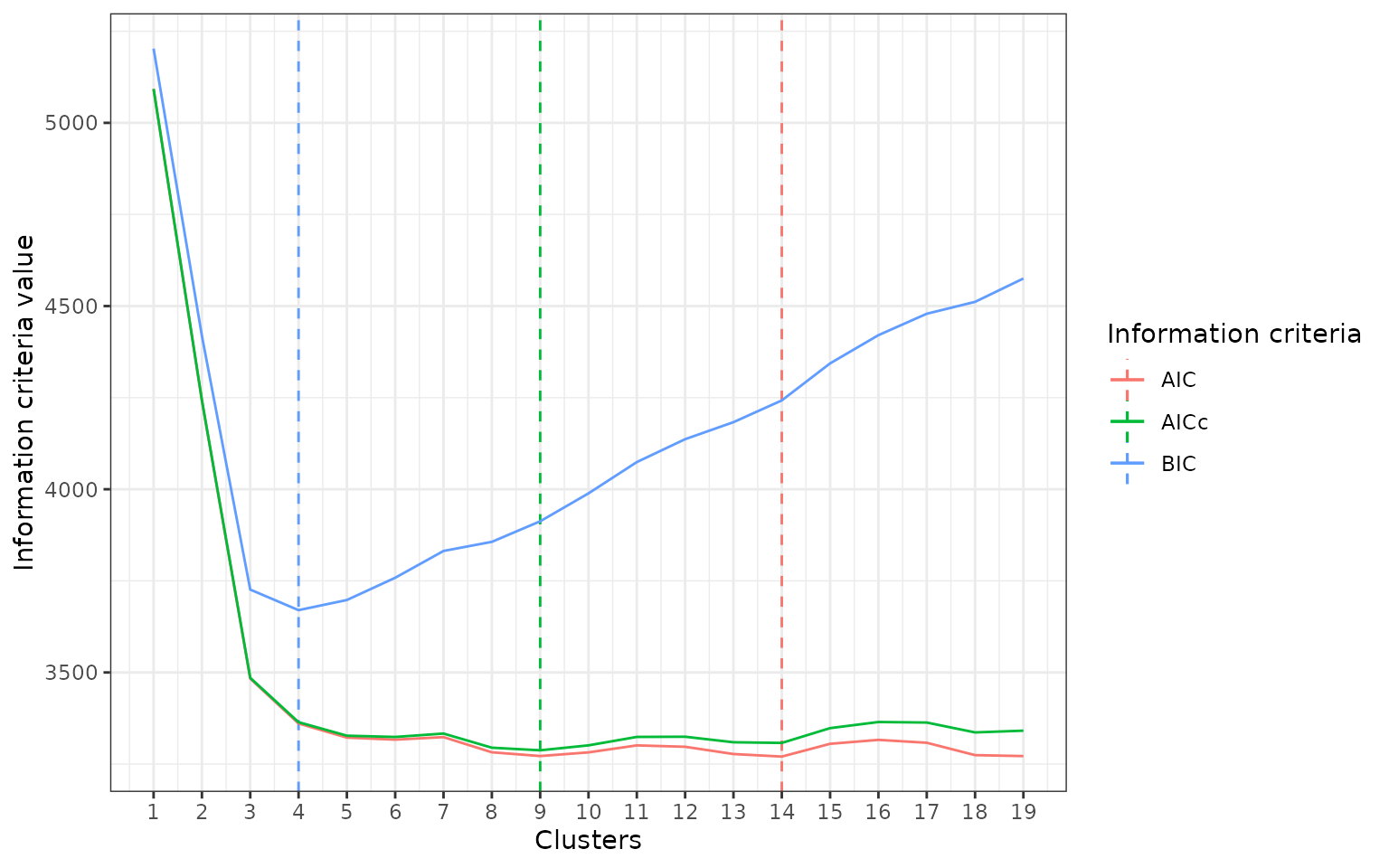
fits <- disclapmix_adaptive(db, criteria = c("BIC", "AIC", "AICc"), margin = 5L)
length(fits)
#> [1] 19
d <- data.frame(
BIC = sapply(fits, function(x) x$BIC_marginal),
AIC = sapply(fits, function(x) x$AIC_marginal),
AICc = sapply(fits, function(x) x$AICc_marginal),
Clusters = sapply(fits, function(x) nrow(x$y))
)
best_BIC <- d$Clusters[which.min(d$BIC)]
best_AIC <- d$Clusters[which.min(d$AIC)]
best_AICc <- d$Clusters[which.min(d$AICc)]
library(ggplot2)
ggplot(d) +
geom_vline(aes(xintercept = best_BIC, color = "BIC"), linetype = "dashed") +
geom_vline(aes(xintercept = best_AIC, color = "AIC"), linetype = "dashed") +
geom_vline(aes(xintercept = best_AICc, color = "AICc"), linetype = "dashed") +
geom_line(aes(Clusters, BIC, color = "BIC")) +
geom_line(aes(Clusters, AIC, color = "AIC")) +
geom_line(aes(Clusters, AICc, color = "AICc")) +
scale_x_continuous(breaks = unique(d$Clusters)) +
labs(y = "Information criteria value", color = "Information criteria") +
theme_bw()